Lipid–protein interactions are central to maintaining the structural and functional balance of biological membranes, influencing a wide array of cellular processes. These interactions, however, become pathological in neurodegenerative diseases (NDDs), such as Alzheimer’s, Parkinson’s, and Huntington’s diseases. In these disorders, the misfolding and aggregation of proteins like amyloid-beta (Aβ), alphasynuclein (α-syn), and mutant huntingtin (mHTT) disrupt the lipid bilayer, compromising membrane integrity, fluidity, and signaling. In this review we explore the critical role of lipid–protein interactions in NDDs, emphasizing how protein misfolding leads to toxic aggregates that embed into membranes, triggering neurotoxic events. Advanced spectroscopic techniques have been instrumental in studying these molecular interactions. Photon-based methods, including Förster resonance energy transfer (FRET), circular dichroism (CD), and Raman spectroscopy, provide real-time insights into protein aggregation and lipid membrane dynamics. Neutron-based techniques, such as neutron reflectometry and small-angle neutron scattering (SANS), further enhance the resolution of lipid–protein interactions, particularly in the context of neurodegenerative aggregation.
Moreover, the review highlights the significance of lipid microdomains, particularly cholesterol-rich lipid rafts, which act as platforms for protein aggregation, influencing disease progression. Therapeutic strategies aimed at targeting these lipid–protein interfaces are also discussed, with a focus on how spectroscopic insights have driven the development of drugs that stabilize membrane integrity or prevent toxic aggregation. Finally, the integration of spectroscopy with computational models, such as molecular dynamics (MD) simulations, is proposed as a promising approach to further unravel the complex dynamics of lipid–protein interactions, providing a more complete picture of disease mechanisms.
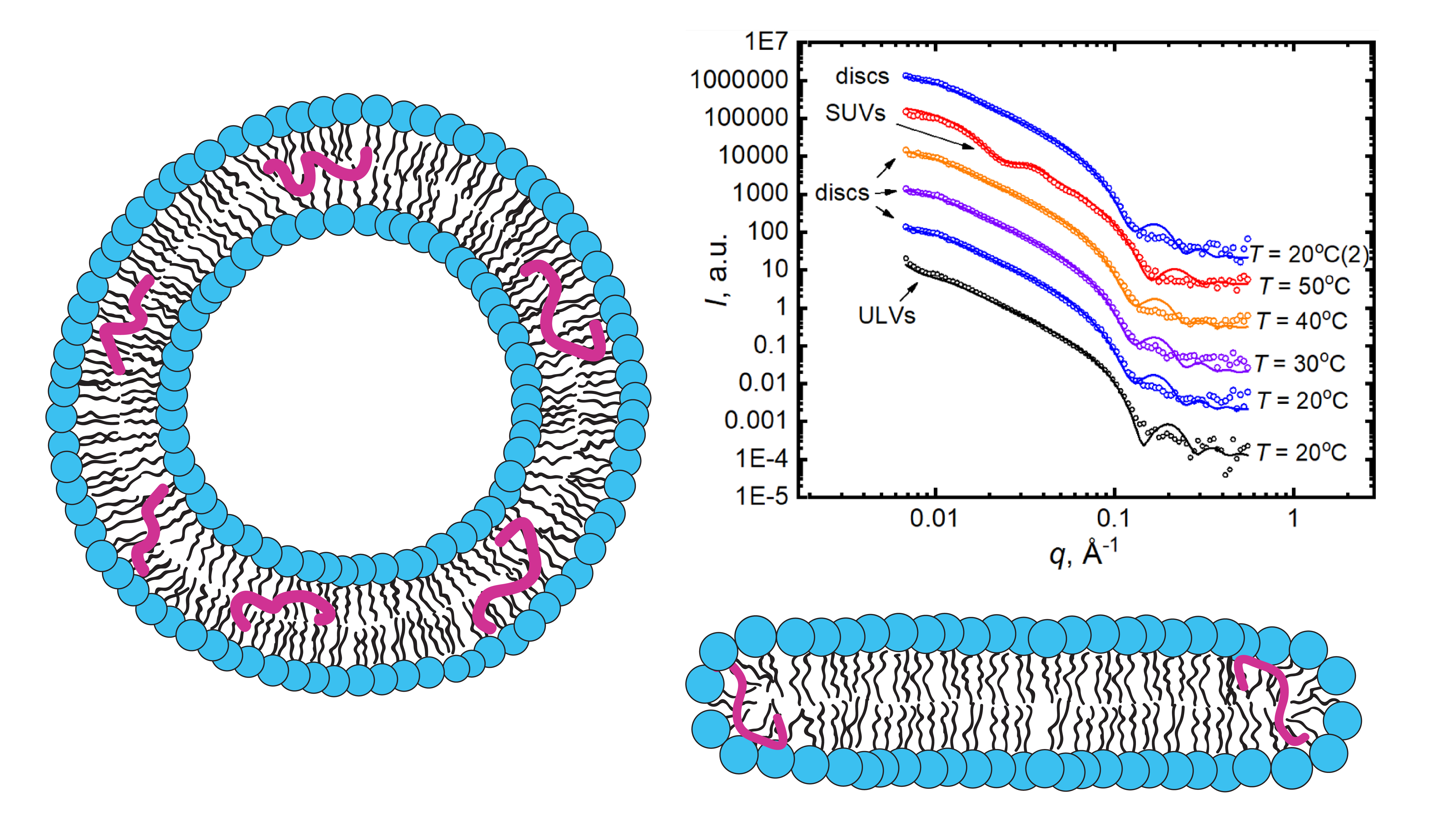
The amyloid-beta peptide (Aβ peptide) is proposed to play a central role in the onset of Alzheimer’s disease (AD). The pathology is associated with the fast accumulation of neurotoxic amyloid aggregates in brain tissues, though the fundamentals of the disease’s progression remain unsolved. It is noted that the preclinical stage of AD may play a crucial role in its further irreversible development. Namely, interactions between lipid membranes and Aβ-peptide molecules incorporated therein at relatively low concentrations should be under a close attention. In this review, we discuss recent works devoted to studying the lipid peptide interactions with a specific focus on the lipid membrane reorganizations caused by Aβ (25–35) peptide in the preclinical AD mimicking conditions. The interactions observed are believed to be important in understanding the mechanisms of the Aβ-peptide destructive effects on lipid membranes and the corresponding onset of the disease. The methods of applied nuclear physics have proven remarkably relevant in such research. The scattering methods provided instrumental information on a level of supramolecular assemblies, while spectrometry allowed obtaining information on the molecular level. Finally, molecular dynamics simulations provided details unachievable by experimental approaches, though the validation role of the latter cannot be undermined. Altogether, the recent advances in research results prove these complementary approaches the most appropriate for tackling the complex issues of biomembrane interactions.
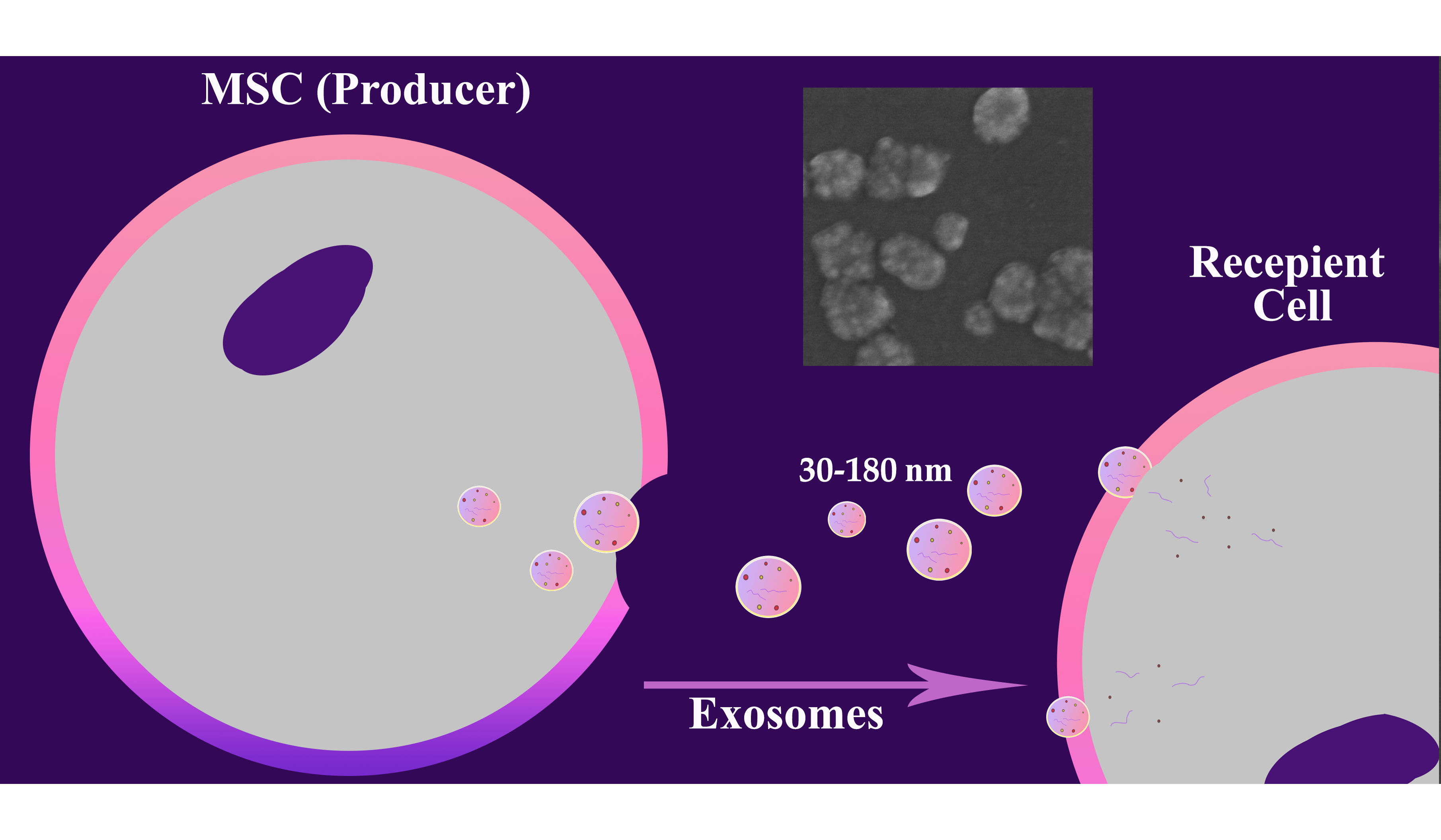
This study provides a comparative analysis of various components of mesenchymal stem cells (MSC) conditioned media (CM) obtained using serum-containing and serum-free culture methods, revealing significant differences in their composition and potential clinical applicability. Serum-containing CM exhibits significantly higher levels of total protein, non-vesicular RNA, exosomes, and nanoparticles compared to serum-free CM, reflecting the contribution of both the MSC secretome and residual fetal bovine serum components. Ultrafiltration-based fractionation (0.2 µm–50 kDa) allows the isolation of fraction enriched in exosomes and proteins, preserving the functionally significant components of the MSC secretome. This strategy effectively captures small vesicles and mid-sized proteins while excluding larger or smaller biomolecules, enhancing utility for targeted analyses. The presented data underscore the need for context-driven CM selection and provide information for choosing the optimal strategy for obtaining the MSC secretome balancing yield, purity, and regulatory demands in MSC research and therapy.
In this work, the characteristics of a prototype SPECT system based on the Timepix readout chip, with a MURA-type encoding mask, were evaluated. The setup has a small FoV and can be used in preclinical studies of drugs on small laboratory animals. Despite many existing test protocols developed and described in pertinent documents of national standard bodies and IAEA recommendations, they are not suitable for microtomographic systems based on semiconductor pixel detectors due to different detector technology, high spatial resolution and small area of interest. To measure their characteristics, special phantoms were developed, with a small “hot region”.
Such micro-SPECT parameters as spatial resolution, contrast, linearity, and system efficiency were studied using 99mTc source. The detector calibration and data preprocessing are described.